Spatial receptive-field of nonlagged cells in dLGN of cat¶
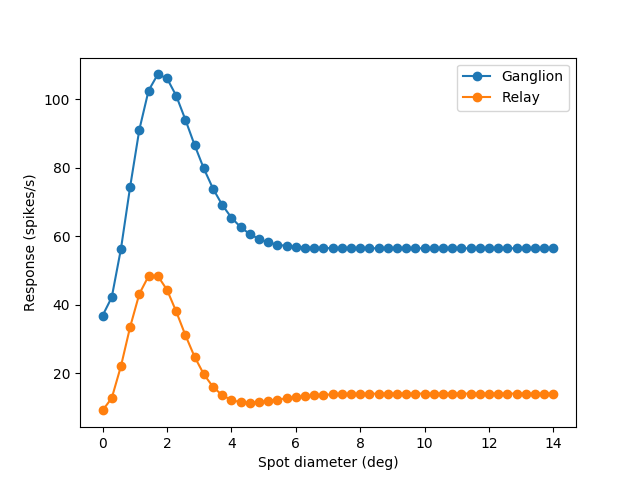
In this example figure 5 in Einevoll et al. (2000) is reproduced. In the code below the size tuning curve for the ganglion cell and the relay cell is calculated. The resulting figure shown below:
import numpy as np
import quantities as pq
import matplotlib.pyplot as plt
import pylgn
import pylgn.kernels as kernel
patch_diameter = np.linspace(0, 14, 50) * pq.deg
R_g = np.zeros(len(patch_diameter)) / pq.s
R_r = np.zeros(len(patch_diameter)) / pq.s
# create network
network = pylgn.Network()
# create integrator
integrator = network.create_integrator(nt=1, nr=8, dt=1*pq.ms, dr=0.1*pq.deg)
# create neurons
ganglion = network.create_ganglion_cell(background_response=36.8/pq.s)
relay = network.create_relay_cell(background_response=9.1/pq.s)
# create kernels
Wg_r = kernel.spatial.create_dog_ft(A=-1, a=0.62*pq.deg, B=-0.85, b=1.26*pq.deg)
Krig_r = kernel.spatial.create_gauss_ft(A=1, a=0.88*pq.deg)
Krg_r = kernel.spatial.create_delta_ft()
# connect neurons
ganglion.set_kernel((Wg_r, kernel.temporal.create_delta_ft()))
network.connect(ganglion, relay, (Krg_r, kernel.temporal.create_delta_ft()), weight=0.81)
network.connect(ganglion, relay, (Krig_r, kernel.temporal.create_delta_ft()), weight=-0.56)
for i, d in enumerate(patch_diameter):
# create stimulus
stimulus = pylgn.stimulus.create_patch_grating_ft(patch_diameter=d, contrast=-131.3)
network.set_stimulus(stimulus)
# compute
network.compute_response(ganglion, recompute_ft=True)
network.compute_response(relay, recompute_ft=True)
R_g[i] = ganglion.center_response[0]
R_r[i] = relay.center_response[0]
# visualize
plt.plot(patch_diameter, R_g, '-o', label="Ganglion")
plt.plot(patch_diameter, R_r, '-o', label="Relay")
plt.xlabel("Spot diameter (deg)")
plt.ylabel("Response (spikes/s)")
plt.legend()
plt.show()